
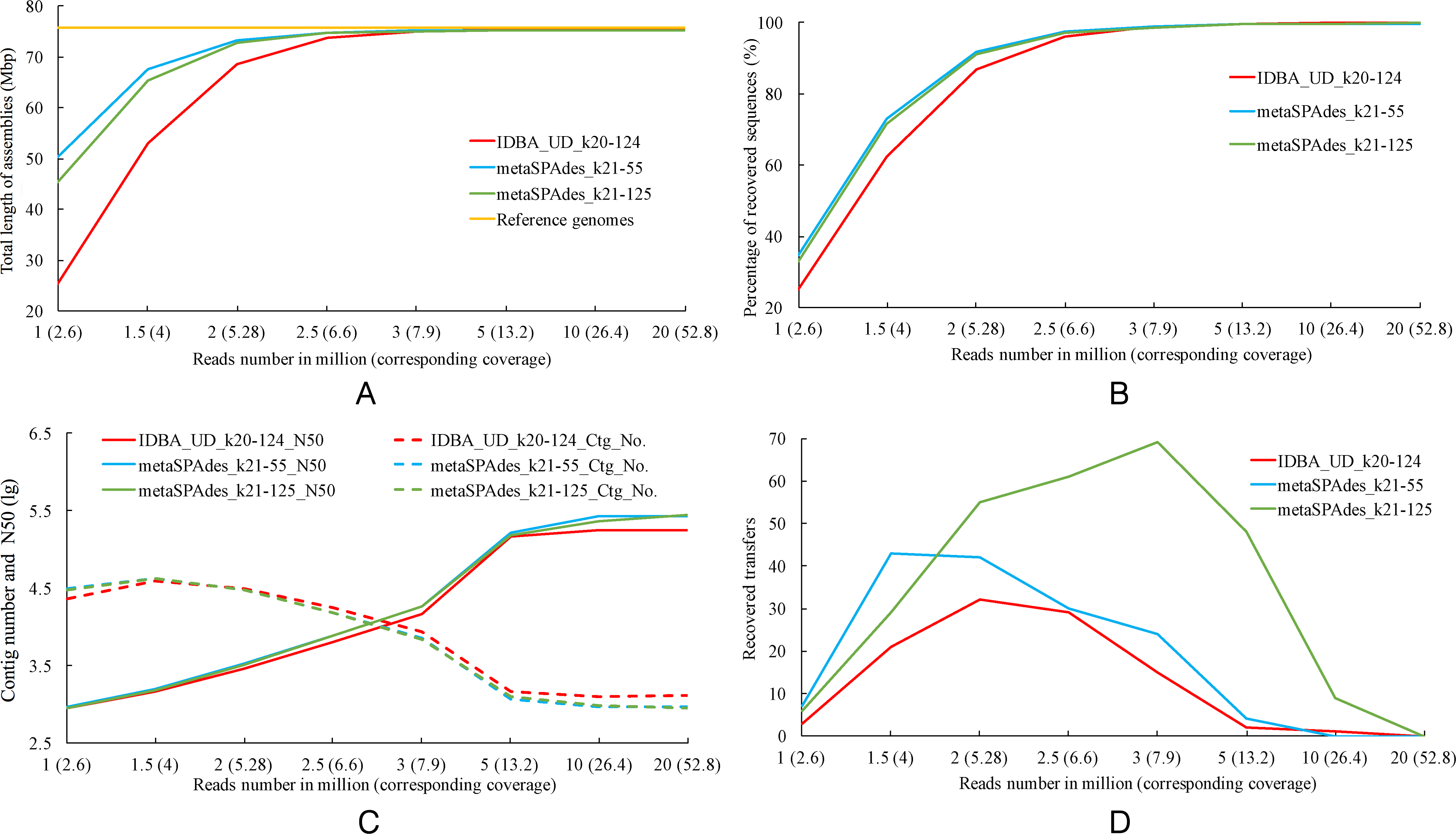
We recommend long-read technologies for any genome with non-trivial repeat content. Their relatively low accuracy and high frequency of small indels makes it desirable to perform polishing and error correction for finished genomes, but Hi-C scaffolding frequently works well even with unpolished contigs. Long read assemblies are strongly encouraged, as they lead to extremely contiguous assemblies due to their ability to completely read through long repetitive sequences. The following sequencing technologies are discouraged for generating starting contigs or scaffolds: Illumina short reads (very high coverage).Sanger / dideoxy sequencing (high coverage).The following sequencing technologies are acceptable in some cases for generating starting contigs or scaffolds: Nanopore long reads (minimum 50X recommend error correction, less well investigated than PacBio).PacBio long reads (ideally >70X coverage minimum 30X).The following molecular technologies are strongly encouraged for generating starting contigs or scaffolds: 1 Assembly technologies Recommendations on sequencing technology for initial (contig-level) assembly If you want a diploid or polyploid assembly please email to learn more about our recommendations. Diploid or polyploid Hi-C scaffolding is possible but more complex. These recommendations cover the most common case of using Hi-C to scaffold a haploid genome assembly. It is therefore important to ensure that your starting assembly is of sufficient quality for the scaffolding task.
Hi-C is a great tool for genome scaffolding, but its success depends crucially on the starting assembly.


 0 kommentar(er)
0 kommentar(er)
